Nice to meet you!
I build healthcare software at Palantir. Before that, I received my PhD in computational genetics at Vanderbilt.
I've built tools that power analysis in DNA biobanks, alternative asset markets, and more.
🚧 🚧 🚧
This website is actively under construction. Check back later for more.
Want to get in touch? Connect with me on LinkedIn or send me an email!
Past Research
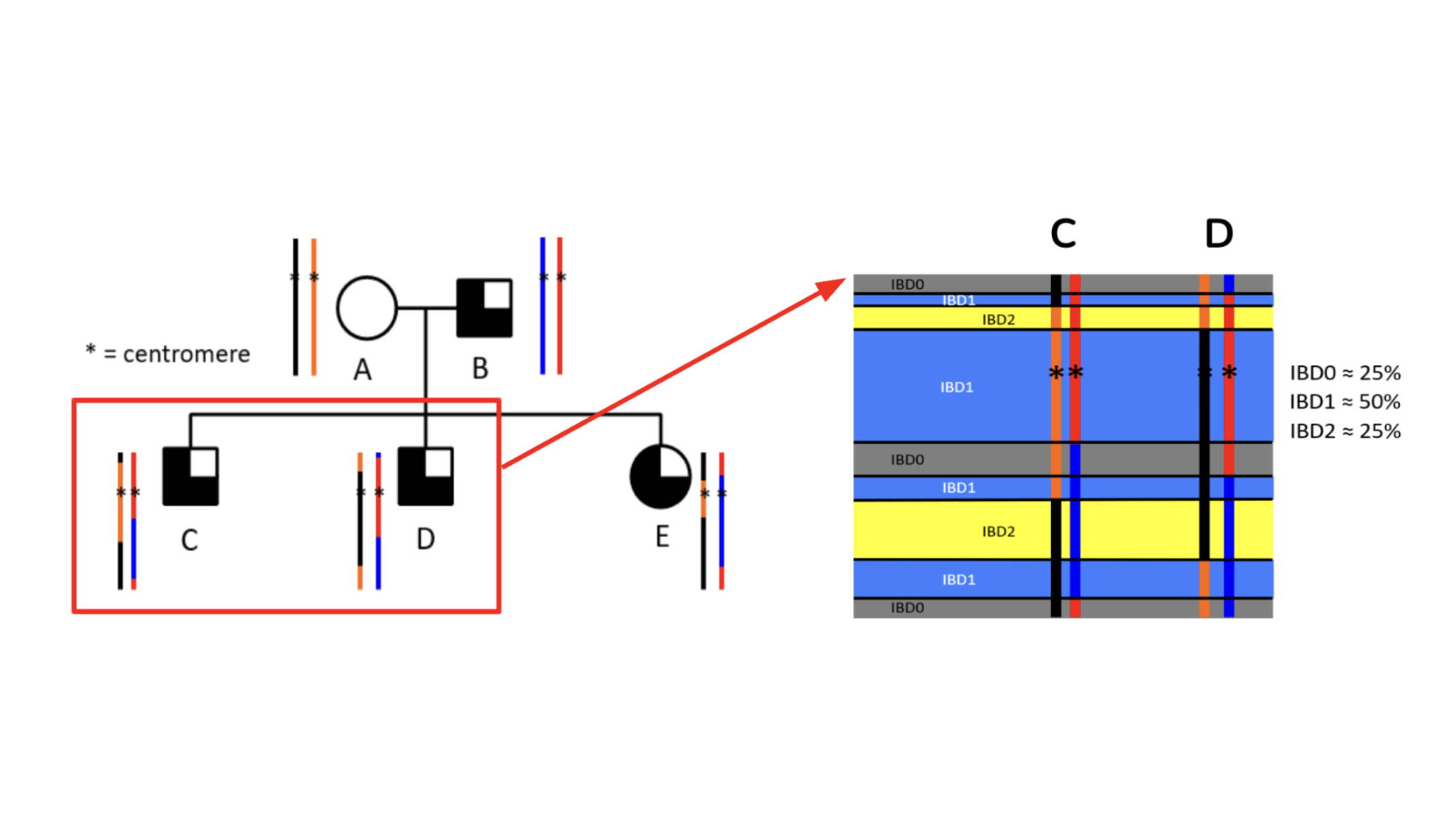
My doctoral work in the
Below Lab at Vanderbilt University was in genetic relatedness analysis.
I developed software
that leverages biobank-scale genetic data to characterize
novel disease associations, build pedigrees out of raw data, and more.
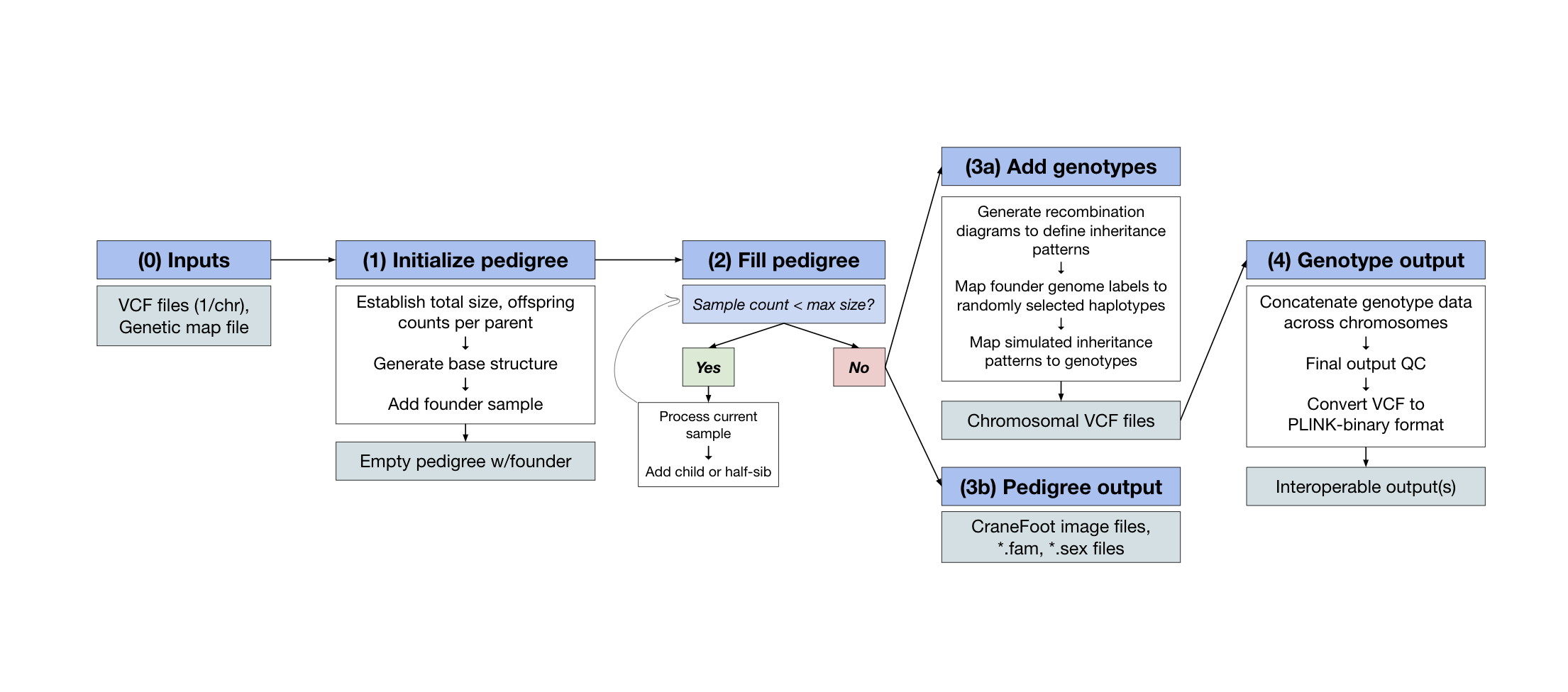
COMPADRE
Pedigree generation software that incorporates global sharing proportions from PRIMUS and segment length & distribution from ERSA.
Built with: Python, Perl, PLINK, Docker
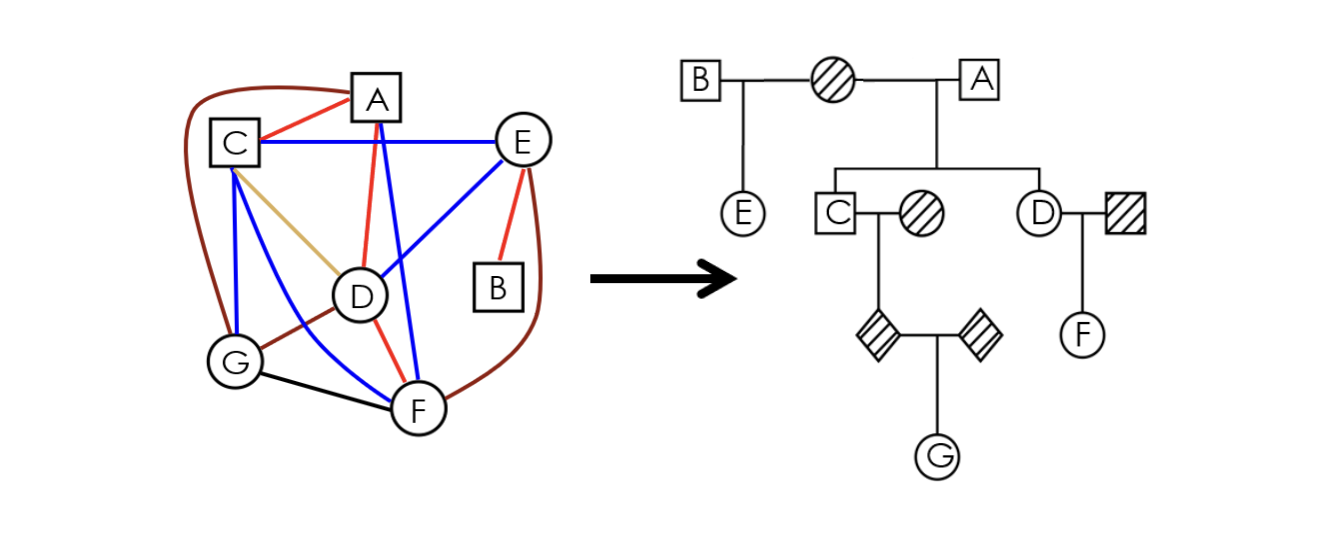
Scalable map-reduce approach to identifying IBD segment-based enrichment of binary trait disease status in large datasets.
Built with: C++, Python, R, SQLite
Education
Vanderbilt University School of Medicine | Ph.D., Human Genetics | 2019-2025
↳ Teaching Assistant, Python (2021-2022)
↳ Award Recipient, Big Biomedical Data Science Training Program (2020-2021)
↳ Award Recipient, Dean's Award (2019-2020)
Duke University | B.S., Biology with Distinction | Class of 2019
↳ Event Coordinator, Duke Medical Entrepreneurship (2016)
↳ Dean's List (2017, 2019)
Publications
I have published three manuscripts as a first author, two as a supporting author, and contributed to dozens of
conference abstracts and presentations.
More information is available on my
Google Scholar profile.